
We further show that the identified CMA substrates display high expression in multiple primary cancers compared to their normal counterparts. In particular, several proteins of the translation initiation complex were identified as bona fide CMA substrates in multiple cancer cell lines of distinct origin and we show that CMA suppresses cellular translation. Beyond verifying metabolic pathways, we show that multiple components involved in select biological processes, including cellular translation, was specifically targeted for degradation by CMA. By integrating bioinformatics analyses, we identified and categorized proteins of multiple cellular pathways that were specifically targeted by CMA. Here, we undertook a quantitative multiplex mass spectrometry approach to study the proteome of isolated lysosomes in cancer cells during CMA-activated conditions. Nearly one-third of the soluble proteins are predicted to be recognized by this pathway, yet only a minor fraction of this proteome has been identified as CMA substrates in cancer cells.
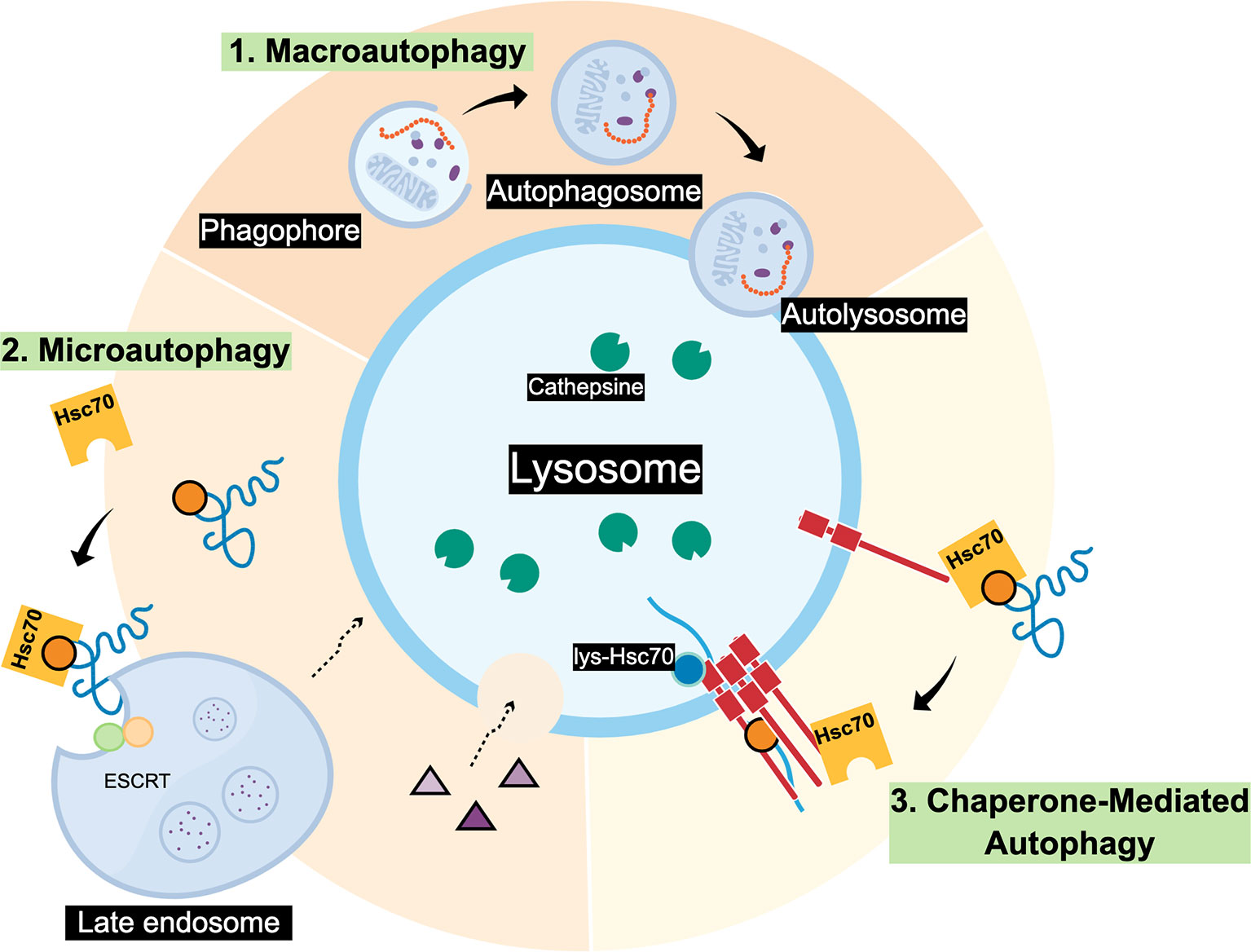
Chaperone-mediated autophagy (CMA) is a lysosomal degradation pathway of select soluble proteins.


 0 kommentar(er)
0 kommentar(er)
